 Author
Author  Correspondence author
Correspondence author
International Journal of Molecular Medical Science, 2024, Vol. 14, No. 4
Received: 03 Jun., 2024 Accepted: 07 Jul., 2024 Published: 19 Jul., 2024
The advent of genomic studies has significantly advanced our understanding of depression and its treatment, paving the way for personalized medicine. This review explores the integration of pharmacogenomics and other omic technologies in the treatment of depression, highlighting the potential for individualized care. Pharmacogenomic studies have identified several single nucleotide polymorphisms (SNPs) that influence the efficacy of antidepressants and mood stabilizers, although their clinical application remains limited. Genome-wide association studies have further elucidated genetic predictors of treatment-resistant depression, suggesting potential biomarkers for personalized treatment. Despite these advancements, the translation of genetic findings into clinical practice has been slow, with current diagnostic and treatment strategies still largely symptom-based. However, the use of pharmacogenomic testing has shown promise in improving treatment outcomes by guiding medication selection. Future research should focus on integrating multi-omic data to develop comprehensive predictive models for treatment response, ultimately enhancing the precision of depression management.
1 Introduction
Depression, clinically known as Major Depressive Disorder (MDD), is a pervasive mood disorder characterized by persistent feelings of sadness, hopelessness, and a lack of interest or pleasure in daily activities. It is a significant public health concern, affecting individuals across all age groups, including adults, teenagers, and children (Sadeeqa, 2018). The World Health Organization estimates that over 300 million people globally suffer from depression, making it the leading cause of disability worldwide (Neto et al., 2019). The impact of depression extends beyond the individual, affecting families, workplaces, and society at large. It is associated with considerable morbidity, mortality, and economic costs, including heightened risks of suicide (Hyde et al., 2016; Wray et al., 2017).
Traditional treatment approaches for depression, which typically include pharmacotherapy and psychotherapy, often yield limited success due to the heterogeneous nature of the disorder (Greden et al., 2019). The variability in treatment response necessitates a more personalized approach to effectively manage and treat depression. Personalized treatment aims to tailor therapeutic strategies to the individual characteristics of each patient, thereby improving treatment outcomes and reducing the trial-and-error process associated with conventional methods (Sekaran and Shanmugam, 2021). This approach is particularly crucial given the complex interplay of genetic, environmental, and psychosocial factors that contribute to the onset and progression of depression (Belmaker and Agam, 2008).
Genomic studies have significantly advanced our understanding of the biological underpinnings of depression. Genome-wide association studies (GWAS) have identified numerous genetic variants associated with depression, highlighting the polygenic nature of the disorder (Howard et al., 2019). For instance, a meta-analysis involving over 800,000 individuals identified 102 independent variants and 269 genes associated with depression, underscoring the importance of genetic factors in its etiology (Howard et al., 2019). These findings pave the way for the development of pharmacogenomic-guided treatments, which use genetic information to predict individual responses to medications and tailor treatment plans accordingly (Greden et al., 2019). Such approaches have shown promise in improving response and remission rates in patients with difficult-to-treat depression (Greden et al., 2019).
This study aims to review the current state of genomic research in the context of depression and explore its implications for personalized treatment. By synthesizing findings from recent genomic studies, this review aims to provide a comprehensive overview of the genetic architecture of depression and discuss how these insights can be translated into clinical practice to enhance treatment efficacy. The ultimate goal is to highlight the potential of genomics to revolutionize the management of depression, offering hope for more effective and individualized therapeutic strategies.
2 Genomic Factors in Depression
2.1 Genetic variants associated with depression
Genome-wide association studies (GWAS) have significantly advanced our understanding of the genetic underpinnings of major depressive disorder (MDD). Recent meta-analyses combining data from multiple large-scale studies have identified several genetic loci associated with MDD. For instance, a meta-analysis involving 90 150 MDD cases and 246 603 controls identified significant associations at loci 6q16.2, 12q24.31, and 16p13.3, suggesting that genetic variants at these loci may confer risk for MDD through the regulation of gene expression in brain tissues (Li et al., 2018). Another study identified 15 genetic loci associated with MDD in individuals of European descent, highlighting the potential of large-scale consumer genomic data to complement traditional research methods (Hyde et al., 2016). These findings underscore the polygenic nature of depression, where multiple genetic variants each contribute a small effect to the overall risk (Cohen-Woods et al., 2012; Shadrina et al., 2018).
Specific genes and loci have been implicated in the risk for depression through GWAS. For example, the FBXL4 and RSRC1 genes located at the 16p13.3 locus were found to be significantly upregulated in the brains of MDD patients, suggesting their involvement in the pathogenesis of depression (Alladi et al., 2018). Additionally, the HACE1 and SHANK2 genes have been implicated through studies examining the interplay between genetic variants and DNA methylation. Lower DNA methylation at the HACE1 promoter was associated with higher risk for depression, while SHANK2 was linked to neuronal processes through miRNA regulation (Ciuculete et al., 2020). These findings provide insights into the molecular mechanisms underlying depression and highlight potential targets for therapeutic intervention.
2.2 Epigenetic factors
Epigenetic modifications, such as DNA methylation and histone modification, play a crucial role in the regulation of gene expression and have been implicated in the pathophysiology of depression. DNA methylation, in particular, has been extensively studied as a potential biomarker for depression. For instance, a meta-analysis of epigenome-wide association studies (EWAS) identified three CpG sites significantly associated with depressive symptoms, pointing towards axon guidance as a disrupted pathway in depression (Jovanova et al., 2018). Histone modifications, although less studied in the context of depression, are known to influence chromatin structure and gene expression, potentially contributing to the development of depressive disorders (Shadrina et al., 2018).
Environmental factors, such as stress and trauma, can induce epigenetic changes that influence the risk of developing depression. Twin studies have shown that monozygotic twins discordant for depression exhibit differences in DNA methylation, suggesting that environmental factors contribute to these epigenetic modifications (Alladi et al., 2018). Additionally, childhood trauma has been linked to alterations in DNA methylation patterns, which may predispose individuals to depression later in life (Li et al., 2018). These findings highlight the importance of considering both genetic and environmental factors in understanding the etiology of depression.
2.3 Gene-environment interactions
The interaction between genetic predisposition and environmental stressors is a critical factor in the development of depression. Studies have shown that individuals with certain genetic variants are more susceptible to the effects of environmental stressors, such as trauma or chronic stress. For example, genetic variants that influence DNA methylation and miRNA levels can modulate an individual's vulnerability to depression in response to environmental factors (Ciuculete et al., 2020). This gene-environment interplay underscores the complexity of depression and the need for a multifaceted approach to its study and treatment (Cohen-Woods et al., 2012; Shadrina et al., 2018).
Several significant gene-environment interactions have been identified in the context of depression. One notable example is the interaction between the serotonin transporter gene (5-HTTLPR) and stressful life events, where individuals with the short allele of 5-HTTLPR are more likely to develop depression following exposure to stress (Shadrina et al., 2018). Another example involves the interaction between childhood trauma and DNA methylation changes, which has been shown to increase the risk of depression in affected individuals (Palma-Gudie et al., 2020). These interactions highlight the importance of considering both genetic and environmental factors in the study of depression and suggest potential avenues for personalized treatment strategies.
In conclusion, the study of genomic factors in depression, including genetic variants, epigenetic modifications, and gene-environment interactions, provides valuable insights into the complex etiology of this disorder. Continued research in these areas holds promise for the development of personalized treatment approaches that take into account an individual's unique genetic and environmental background.
3 Individualized Treatment Strategies for Depression
3.1 Pharmacogenomics
Pharmacogenomics is the study of how genes affect a person's response to drugs. This field combines pharmacology and genomics to develop effective, safe medications and doses tailored to a person's genetic makeup. Genetic differences can significantly influence drug metabolism and response, impacting the efficacy and safety of antidepressants. For instance, variations in genes encoding cytochrome P450 enzymes (such as CYP2D6 and CYP2C19) can alter the metabolism of many psychotropic medications, leading to differences in drug levels and therapeutic outcomes among individuals (Amare et al., 2017; Westrhenen et al., 2020). These genetic differences can result in some patients experiencing adverse effects or inadequate therapeutic responses, necessitating adjustments in medication type or dosage (Murphy and McMahon, 2013).
Several pharmacogenomic markers have been identified that influence the response to antidepressants. For example, single nucleotide polymorphisms (SNPs) in genes such as COMT, HTR2A, and SLC6A4 have been associated with variations in response to selective serotonin reuptake inhibitors (SSRIs) (Amare et al., 2017). The GUIDED trial demonstrated that patients whose treatment was guided by pharmacogenomic testing showed significant improvements in response and remission rates compared to those receiving treatment as usual (Greden et al., 2019). Additionally, combinatorial pharmacogenomic tests, which analyze multiple genetic markers, have been shown to improve clinical outcomes by guiding medication selection and dosing (Tuson et al., 2017; Thase et al., 2019). These tests can categorize medications based on their compatibility with a patient's genetic profile, helping clinicians make more informed treatment decisions.
3.2 Personalized psychotherapy
Emerging evidence suggests that genetic information can also be used to personalize psychotherapy. For instance, genetic variations in the serotonin transporter gene (SLC6A4) have been linked to differential responses to cognitive-behavioral therapy (CBT) (Amare et al., 2017). Studies have shown that individuals with certain genetic profiles may benefit more from specific types of psychotherapy, indicating that genetic information can help tailor therapeutic approaches to individual patients (Amare et al., 2017). This personalized approach aims to enhance the effectiveness of psychotherapy by aligning treatment strategies with the patient's genetic predispositions.
Specific therapeutic approaches can be informed by genetic data to optimize treatment outcomes. For example, patients with certain genetic markers associated with better responses to CBT may be prioritized for this type of therapy, while those with markers indicating a poor response might be directed towards alternative therapies such as interpersonal therapy (IPT) or pharmacotherapy (Amare et al., 2017). Additionally, integrating genetic information with other clinical data can help develop comprehensive treatment plans that address the unique needs of each patient, potentially improving adherence and outcomes (Figure 1) (Amare et al., 2017).
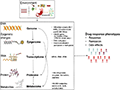 Figure 1 Multilayered Biological Interactions Influencing Drug-Response Phenotypes: From Genome to Metabolome (Adopted from Amare et al., 2017) Image caption: An overview of the biological data that need to be integrated into the systems genomics approach to investigate pathways in complex traits (e.g., treatment response) (Adopted from Amare et al., 2017) |
3.3 Integrative treatment models
Integrative treatment models that combine pharmacogenomics, personalized psychotherapy, and lifestyle interventions hold promise for improving outcomes in depression treatment. By leveraging genetic information, clinicians can tailor both pharmacological and psychotherapeutic interventions to the individual, while also incorporating lifestyle modifications such as diet, exercise, and sleep hygiene to support overall mental health (Amare et al., 2017; Cheng et al., 2023). This holistic approach aims to address the multifaceted nature of depression, providing a more comprehensive and personalized treatment strategy.
Several studies have demonstrated the effectiveness of integrative treatment models. For instance, the GUIDED trial showed that pharmacogenomic-guided treatment significantly improved response and remission rates in patients with major depressive disorder (MDD) (Figure 2) (Greden et al., 2019). Another study highlighted the potential of combining pharmacogenomic data with personalized psychotherapy and lifestyle interventions to enhance treatment outcomes (Amare et al., 2017). These integrative models have been shown to be particularly beneficial for patients with treatment-resistant depression, where traditional approaches have failed (Cheng et al., 2023). Further research and clinical trials are needed to refine these models and establish their efficacy across diverse patient populations.
 Figure 2 Patient outcomes at week 8 in the pharmacogenomics guided-care arm (n = 560) compared to treatment as usual (n = 607) (Adopted from Greden et al., 2019) Image caption: Outcomes were evaluated using the HAM-D17 depression rating scales (Adopted from Greden et al., 2019) |
In conclusion, individualized treatment strategies for depression, including pharmacogenomics, personalized psychotherapy, and integrative treatment models, offer promising avenues for improving patient outcomes. By tailoring interventions to the genetic and clinical profiles of patients, these approaches aim to enhance the efficacy and safety of depression treatments, ultimately leading to better mental health and quality of life for individuals affected by this condition.
4 Impact of Genomics on Treatment Outcomes
4.1 Efficacy of personalized treatments
The comparative effectiveness of personalized treatments versus standard treatments has been a focal point in recent research. Pharmacogenomics, which tailors medication based on genetic profiles, has shown promise in improving treatment outcomes for major depressive disorder (MDD). For instance, the GUIDED trial demonstrated that while symptom improvement was not significantly different between pharmacogenomics-guided care and treatment as usual (TAU), there were significant improvements in response and remission rates for patients receiving guided care (Greden et al., 2019). Similarly, a meta-analysis of randomized controlled trials (RCTs) found that pharmacogenomics-guided treatment was superior to TAU in terms of response and remission rates for treatment-resistant depression (TRD) (Figure 3) (Cheng et al., 2023). These findings suggest that personalized treatments can offer substantial benefits over standard approaches, particularly for patients who have not responded to initial treatments.
 Figure 3 Meta-analysis results of primary outcomes (Adopted from Cheng et al., 2023) Image caption: (A) comparison of the proportion of patients achieving response with guided versus unguided treatment; (B) comparison of the proportion of patients achieving remission with guided versus unguided treatment. CI, confidence interval; PGx, pharmacogenomics; TAU, treatment as usual (Adopted from Cheng et al., 2023) |
Clinical trials have provided concrete examples of the efficacy of personalized treatments. In the IMPACT study, patients with MDD who received combinatorial pharmacogenomic testing showed significant improvements in symptom reduction, response, and remission rates compared to those who did not receive such testing (Tanner et al., 2018). Another study highlighted that patients taking medications congruent with their genetic profiles experienced better outcomes than those on incongruent medications (Thase et al., 2019). These trials underscore the potential of personalized medicine to enhance treatment efficacy by aligning medication choices with individual genetic profiles.
4.2 Patient adherence and satisfaction
Patient adherence and satisfaction are critical components of successful treatment outcomes. Pharmacogenomics-guided treatments have been shown to positively impact these factors. For example, a meta-analysis revealed that while the acceptability and side effect burden of pharmacogenomics-guided treatments were not significantly different from TAU, the guided treatments did show a trend towards better patient adherence (Cheng et al., 2023). This suggests that personalized treatments may enhance patient satisfaction by reducing the trial-and-error process often associated with standard treatments.
Qualitative studies and patient feedback further illuminate the benefits of personalized treatments. Patients often report higher satisfaction levels when their treatment plans are tailored to their genetic profiles. In the GUIDED trial, patients who switched from incongruent to congruent medications reported greater symptom improvement and higher satisfaction with their treatment (Greden et al., 2019). These qualitative insights highlight the importance of considering patient experiences and preferences in the development of personalized treatment plans.
4.3 Cost-effectiveness
Economic evaluations have shown that personalized treatment approaches can be cost-effective. The initial costs of pharmacogenomic testing are often offset by the reduction in ineffective treatments and the associated healthcare costs. For instance, a study on the economic impact of pharmacogenomics-guided treatments found that the improved response and remission rates led to lower overall healthcare costs by reducing the need for additional treatments and hospitalizations (Rosenblat et al., 2018). This suggests that personalized treatments not only improve clinical outcomes but also offer economic benefits.
Cost-benefit analyses further support the economic viability of personalized treatments. By improving treatment efficacy and reducing the duration of ineffective treatments, pharmacogenomics-guided care can lead to significant cost savings. For example, the IMPACT study demonstrated that patients receiving pharmacogenomic-guided care had better outcomes and required fewer additional treatments, resulting in lower overall healthcare costs (Thase et al., 2019). These findings highlight the potential for personalized treatments to provide both clinical and economic advantages.
In summary, the impact of genomics on treatment outcomes for depression is multifaceted, encompassing improvements in efficacy, patient adherence, satisfaction, and cost-effectiveness. Personalized treatments, guided by pharmacogenomic testing, offer a promising approach to enhancing the management of depression, particularly for patients who have not responded to standard treatments. The evidence from clinical trials and economic evaluations underscores the potential benefits of integrating genomics into routine clinical practice.
5 Challenges and Limitations
5.1 Ethical and privacy concerns
The integration of genomic data into the treatment of depression raises significant ethical and privacy issues. One of the primary concerns is the potential misuse of genetic information. Patients may fear that their genetic data could be used against them by employers or insurance companies, leading to discrimination. This fear can deter individuals from participating in genomic studies or utilizing pharmacogenomic testing, thereby limiting the potential benefits of personalized treatment approaches (Amareet al., 2017; Fabbri et al., 2018; Greden et al., 2019).
Moreover, the storage and handling of genetic data require stringent security measures to prevent unauthorized access and breaches. Ensuring that patients' genetic information is kept confidential and secure is paramount to maintaining trust in genomic medicine. Ethical guidelines and robust legal frameworks are necessary to protect individuals' privacy and to regulate the use of genetic data in clinical settings (Ormelet al., 2019; Singh et al., 2023).
5.2 Clinical implementation
The clinical implementation of pharmacogenomic testing in the treatment of depression faces several challenges. One significant barrier is the lack of standardized guidelines for the use of genetic information in clinical practice. While some genetic variants have been associated with antidepressant response, there is no consensus on how to incorporate these findings into treatment decisions (Rosenblat et al., 2017; Fabbri et al., 2020).
Additionally, the cost of pharmacogenomic testing can be prohibitive for many patients, and insurance coverage for these tests is often limited. This financial barrier can prevent widespread adoption of personalized treatment approaches. Furthermore, healthcare providers may lack the necessary training and resources to interpret genetic test results and apply them effectively in clinical practice. This gap in knowledge and expertise can hinder the integration of pharmacogenomics into routine care (Rosenblat et al., 2017; Alladi et al., 2018).
5.3 Limitations of current studies and potential biases
Current studies on the use of pharmacogenomics in the treatment of depression have several limitations and potential biases. Many studies have small sample sizes, which can limit the generalizability of the findings. Additionally, the majority of research has focused on specific populations, often excluding diverse ethnic groups. This lack of diversity can result in findings that are not applicable to all patient populations (Fabbri et al., 2018; Greden et al., 2019; Brown et al., 2022).
Another limitation is the variability in study designs and methodologies. Differences in the genetic tests used, the criteria for defining treatment response, and the duration of follow-up can lead to inconsistent results. This heterogeneity makes it challenging to draw definitive conclusions about the efficacy of pharmacogenomic-guided treatment (Amareet al., 2017; Fabbri et al., 2020).
Potential biases in the studies also need to be considered. Industry-sponsored research may have conflicts of interest that could influence the study outcomes. Additionally, publication bias, where positive results are more likely to be published than negative or null findings, can skew the evidence base. Addressing these biases through rigorous study designs, transparent reporting, and independent replication of findings is essential for advancing the field (Rosenblat et al., 2017; 2018).
In conclusion, while genomic studies hold promise for personalized treatment of depression, several challenges and limitations must be addressed. Ethical and privacy concerns, barriers to clinical implementation, and limitations of current studies highlight the need for continued research, standardized guidelines, and robust ethical frameworks to fully realize the potential of pharmacogenomics in improving treatment outcomes for depression.
6 Future Directions
6.1 Advances in genomic technologies
The field of genomics has seen significant advancements in recent years, which hold promise for the personalized treatment of depression. Genome-wide association studies (GWAS) have identified numerous loci associated with depression, providing a foundation for further exploration of the genetic underpinnings of the disorder (Ormel et al., 2019). However, the challenge remains in identifying the causal mechanisms behind these associations, as GWAS results often highlight genomic regions without direct links to biological functions (Ormel et al., 2019). Future research should focus on integrating data from various omic pillars, such as genomics, epigenomics, proteomics, and metabolomics, to create a comprehensive understanding of the biological pathways involved in depression and its treatment (Amare et al., 2017).
Additionally, advances in DNA methylation studies have shown potential in identifying biomarkers for treatment response in major depressive disorder (MDD) (Alladi et al., 2018). These epigenetic modifications can provide insights into gene-environment interactions that influence both the pathophysiology and treatment response of depression. As genomic technologies continue to evolve, the development of more sophisticated tools for data integration and analysis will be crucial in advancing personalized medicine in psychiatry (Amare et al., 2017; Alladi et al., 2018).
6.2 Expanding personalized treatment
Personalized treatment for depression aims to tailor therapeutic interventions based on an individual's genetic makeup, clinical characteristics, and other biomarkers. Pharmacogenomic studies have already identified several genetic variants that influence the efficacy of antidepressants and mood stabilizers, such as selective serotonin reuptake inhibitors (SSRIs) and lithium (Amare et al., 2017). These findings have led to the development of genotyping tests that can guide medication selection, improving treatment outcomes by identifying responders and non-responders, and minimizing adverse drug events (Miller and O'Callaghan, 2013).
Despite these advancements, the clinical utility of genetic biomarkers for personalized treatment remains limited. Future research should focus on improving the design of pharmacogenomic studies by incorporating larger cohorts, more comprehensive genetic analyses, and the integration of clinical-demographic predictors (Fabbri et al., 2018). Additionally, the development of multivariable diagnostic or prognostic algorithms that combine genomic information with other predictors, such as neuroimaging and clinical characteristics, could enhance the accuracy of treatment predictions (Amare et al., 2017).
The use of deep learning and other advanced computational techniques also holds promise for predicting antidepressant response and remission. By analyzing genetic and clinical factors, these models can identify complex relationships between biomarkers and treatment outcomes, potentially leading to more effective personalized treatment strategies (Lin et al., 2018).
6.3 Interdisciplinary research
The future of personalized treatment for depression lies in interdisciplinary research that combines expertise from various fields, including genomics, psychiatry, bioinformatics, and clinical practice. Collaborative efforts are essential to address the complexities of depression and its treatment, as well as to develop innovative approaches for integrating diverse data types.
One area of interdisciplinary research that shows promise is the integration of pharmacogenomics with other omic data, such as proteomics and metabolomics, to identify novel biomarkers and therapeutic targets (Holsboer, 2008; Amare et al., 2017; Chen et al., 2024). This systems genomics approach aims to understand the biological pathways and networks underlying drug response, ultimately leading to more precise and effective treatments.
Another important aspect of interdisciplinary research is the collaboration between basic and clinical scientists to translate genomic findings into clinical practice. This includes the development of diagnostic tests, therapeutic interventions, and predictive algorithms that can be used by clinicians to guide treatment decisions (Miller and O'Callaghan, 2013; Amare et al., 2017). Additionally, the involvement of bioinformaticians and data scientists is crucial for developing advanced computational tools and models that can handle the complexity of genomic and clinical data (Lin et al., 2018).
Interdisciplinary research should also focus on addressing the ethical, legal, and social implications of personalized medicine. This includes ensuring patient privacy and data security, as well as addressing potential disparities in access to personalized treatments. By fostering collaboration across disciplines, the field of personalized treatment for depression can continue to advance and ultimately improve patient outcomes.
7 Concluding Remarks
The integration of genomic studies into the treatment of depression has shown promising advancements towards personalized medicine. Pharmacogenomic research has identified several single nucleotide polymorphisms (SNPs) that influence the efficacy of antidepressants and mood stabilizers, such as those in genes like COMT, HTR2A, and BDNF. Genome-wide association studies (GWAS) have further highlighted gene sets involved in cyclic adenosine monophosphate mediated signal and chromatin silencing as potential biomarkers for treatment-resistant depression (TRD). Additionally, deep learning models have been developed to predict antidepressant response using genetic and clinical biomarkers, demonstrating significant potential in distinguishing responders from non-responders. However, the translation of these findings into clinical practice remains complex due to the multifaceted nature of gene-environment interactions.
The findings from genomic studies have several implications for clinical practice. The identification of genetic markers associated with antidepressant response can guide clinicians in selecting the most effective treatment for individual patients, potentially reducing the trial-and-error approach currently prevalent in depression treatment. Pharmacogenomic testing, as demonstrated in randomized controlled trials, has shown to improve response and remission rates in patients with major depressive disorder (MDD) by informing medication selection based on genetic profiles. Moreover, the development of multivariable diagnostic algorithms that incorporate genomic data alongside other predictors could further enhance the precision of personalized treatment plans. However, the clinical utility of these approaches requires further validation in diverse populations to ensure their efficacy across different ethnic and genetic backgrounds.
The journey towards personalized treatment of depression through genomic studies is both promising and challenging. While significant strides have been made in identifying genetic markers and developing predictive models, the complexity of depression as a disorder necessitates a multifaceted approach that integrates genomics with other omic data and clinical characteristics. Future research should focus on large-scale, diverse population studies to validate and refine these genomic tools, ensuring their broad applicability and effectiveness. The ultimate goal is to achieve a more predictive, preventive, and personalized approach to psychiatry, improving outcomes for patients with depression.
Acknowledgments
The authors extend sincere thanks to two anonymous peer reviewers for their feedback on the manuscript.
Conflict of Interest Disclosure
The authors affirm that this research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Alladi C., Étain B., Bellivier F., and Marie-Claire C., 2018, DNA methylation as a biomarker of treatment response variability in serious mental illnesses: a systematic review focused on bipolar disorder, schizophrenia, and major depressive disorder, International Journal of Molecular Sciences, 19(10): 3026.
https://doi.org/10.3390/ijms19103026
PMid:30287754 PMCid:PMC6213157
Amare A., Schubert K., and Baune B., 2017, Pharmacogenomics in the treatment of mood disorders: strategies and opportunities for personalized psychiatry, EPMA Journal, 8: 211-227.
https://doi.org/10.1007/s13167-017-0112-8
PMid:29021832 PMCid:PMC5607053
Belmaker R., and Agam G., 2008, Major depressive disorder, The New England Journal of Medicine, 358(1): 55-68.
https://doi.org/10.1056/NEJMra073096
PMid:18172175
Brown L., Stanton J., Bharthi K., Maruf A., Müller D., and Bousman C., 2022, Pharmacogenomic testing and depressive symptom remission: a systematic review and meta‐analysis of prospective, controlled clinical trials, Clinical Pharmacology and Therapeutics, 112: 1303-1317.
https://doi.org/10.1002/cpt.2748
PMid:36111494 PMCid:PMC9827897
Chen S.Y., 2024, Crossing disease boundaries: how ai drives rare disease drug discovery, Bioscience Evidence, 14(1): 21-28.
https://doi.org/10.5376/be.2024.14.0003
Cheng Y., Liu H., Yuan R., Yuan K., and Yu S., 2023, Effectiveness of pharmacogenomics on the response and remission of treatment-resistant depression: a meta-analysis of randomised controlled trials, General Psychiatry, 36(6): 2023.
https://doi.org/10.1136/gpsych-2023-101050
PMid:38155841 PMCid:PMC10753713
Ciuculete D., Voisin S., Kular L., Jonsson J., Rask-Andersen M., Mwinyi J., and Schiöth H., 2020, meQTL and ncRNA functional analyses of 102 GWAS-SNPs associated with depression implicate HACE1 and SHANK2 genes, Clinical Epigenetics, 12: 1-4.
https://doi.org/10.1186/s13148-020-00884-8
PMid:32616021 PMCid:PMC7333393
Cohen-Woods S., Craig I., and McGuffin P., 2012, The current state of play on the molecular genetics of depression, Psychological Medicine, 43: 673-687.
https://doi.org/10.1017/S0033291712001286
PMid:22687339
Fabbri C., Corponi F., Souery D., Kasper S., Montgomery S., Zohar J., Rujescu D., Mendlewicz J., and Serretti A., 2018, The genetics of treatment-resistant depression: a critical review and future perspectives, International Journal of Neuropsychopharmacology, 22: 93-104.
https://doi.org/10.1093/ijnp/pyy024
Greden J., Parikh S., Rothschild A., Thase M., Dunlop B., Debattista C., Conway C., Forester B., Mondimore F., Shelton R., Macaluso M., Li J., Brown K., Gilbert A., Burns L., Jablonski M., and DeChairo B., 2019, Impact of pharmacogenomics on clinical outcomes in major depressive disorder in the GUIDED trial: A large, patient- and rater-blinded, randomized, controlled study, Journal of Psychiatric Research, 111: 59-67.
https://doi.org/10.1016/j.jpsychires.2019.01.003
Holsboer F., 2008, How can we realize the promise of personalized antidepressant medicines? Nature Reviews Neuroscience, 9: 638-646.
https://doi.org/10.1038/nrn2453
Howard D., Adams M., Clarke T., Hafferty J., Gibson J., Shirali M., Coleman J., Hagenaars S., Ward J., Wigmore E., Alloza C., Shen X., Barbu M., Xu E., Whalley H., Marioni R., Porteous D., Davies G., Deary I., Hemani G., Berger K., Teismann H., Rawal R., Arolt V., Baune B., Dannlowski U., Domschke K., Tian C., Hinds D., Trzaskowski M., Byrne E., Ripke S., Smith D., Sullivan P., Wray N., Breen G., Lewis C., and McIntosh A., 2019, Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions, Nature Neuroscience, 22: 343-352.
https://doi.org/10.1038/s41593-018-0326-7
Hyde C., Nagle M., Tian C., Chen X., Paciga S., Wendland J., Tung J., Hinds D., Perlis R., and Winslow A., 2016, Identification of 15 genetic loci associated with risk of major depression in individuals of European descent, Nature Genetics, 48(9): 1031-1036.
https://doi.org/10.1038/ng.3623
Jovanova O., Nedeljkovic I., Spieler D., Walker R., Liu C., Luciano M., Bressler J., Brody J., Drake A., Evans K., Gondalia R., Kunze S., Kuhnel B., Lahti J., Lemaitre R., Marioni R., Swenson B., Himali J., Wu H., Li Y., McRae A., Russ T., Stewart J., Wang Z., Zhang G., Ladwig K., Uitterlinden A., Guo X., Peters A., Räikkönen K., Starr J., Waldenberger M., Wray N., Whitsel E., Sotoodehnia N., Seshadri S., Porteous D., Meurs J., Mosley T., McIntosh A., Mendelson M., Levy D., Hou L., Eriksson J., Fornage M., Deary I., Baccarelli A., Tiemeier H., and Amin N., 2018, DNA methylation signatures of depressive symptoms in middle-aged and elderly persons: meta-analysis of multiethnic epigenome-wide studies, JAMA Psychiatry, 75: 949-959.
https://doi.org/10.1001/jamapsychiatry.2018.1725
PMid:29998287 PMCid:PMC6142917
Li X., Luo Z., Gu C., Hall L., McIntosh A., Zeng Y., Porteous D., Hayward C., Li M., Yao Y., Zhang C., and Luo X., 2018, Common variants on 6q16.2, 12q24.31 and 16p13.3 are associated with major depressive disorder, Neuropsychopharmacology, 43(10): 2146-2153.
https://doi.org/10.1038/s41386-018-0078-9
PMid:29728651 PMCid:PMC6098070
Lin E., Kuo P., Liu Y., Yu Y., Yang A., and Tsai S., 2018, A deep learning approach for predicting antidepressant response in major depression using clinical and genetic biomarkers, Frontiers in Psychiatry, 9: 290.
https://doi.org/10.3389/fpsyt.2018.00290
PMid:30034349 PMCid:PMC6043864
Miller D., and O'Callaghan J., 2013, Personalized medicine in major depressive disorder-opportunities and pitfalls, Metabolism, 62(Suppl 1): S34-S39.
https://doi.org/10.1016/j.metabol.2012.08.021
PMid:23021040 PMCid:PMC4672728
Murphy E., and McMahon F., 2013, Pharmacogenetics of antidepressants, mood stabilizers, and antipsychotics in diverse human populations, Discovery Medicine, 16(87): 113-122.
Neto F., and Rosa J., 2019, Depression biomarkers using non-invasive EEG: a review, Neuroscience and Biobehavioral Reviews, 105: 83-93.
https://doi.org/10.1016/j.neubiorev.2019.07.021
Ormel J., Hartman C., and Snieder H., 2019, The genetics of depression: successful genome-wide association studies introduce new challenges, Translational Psychiatry, 9(1): 114.
https://doi.org/10.1038/s41398-019-0450-5
Palma-Gudiel H., Córdova-Palomera A., Navarro V., and Fañanás L., 2020, Twin study designs as a tool to identify new candidate genes for depression: a systematic review of DNA methylation studies, Neuroscience and Biobehavioral Reviews, 112: 345-352.
Rosenblat J., Lee Y., and McIntyre R., 2017, Does pharmacogenomic testing improve clinical outcomes for major depressive disorder? a systematic review of clinical trials and cost-effectiveness studies, The Journal of Clinical Psychiatry, 78(6): 720-729.
https://doi.org/10.4088/JCP.15r10583
Sadeeqa S., 2018, Depression: A Case Study, Alzheimer's Research and Therapy Open Access, 1(1): 19.
https://doi.org/10.4324/9780203380383-19
Sekaran K., and Shanmugam S., 2021, Predicting drug responsiveness by citalopram induced pathway regulations and biomarker discovery in lymphoblastoid cell lines from depression affected individuals, 2021 International Conference on Decision Aid Sciences and Application (DASA), (2021): 267-271.
https://doi.org/10.1109/DASA53625.2021.9682274
Shadrina M., Bondarenko E., and Slominsky P., 2018, Genetics factors in major depression disease, Frontiers in Psychiatry, 9: 334.
https://doi.org/10.3389/fpsyt.2018.00334
Singh P., Srivastava A., Guin D., Thakran S., Yadav J., Chandna P., Sood M., Chadda R., and Kukreti R., 2023, Genetic landscape of major depressive disorder: assessment of potential diagnostic and antidepressant response markers, International Journal of Neuropsychopharmacology, 26: 692-738.
https://doi.org/10.1093/ijnp/pyad001
Tanner J., Davies P., Voudouris N., Shahmirian A., Herbert D., Braganza N., Gugila A., DeChairo B., and Kennedy J., 2018, Combinatorial pharmacogenomics and improved patient outcomes in depression: Treatment by primary care physicians or psychiatrists, Journal of Psychiatric Research, 104: 157-162.
https://doi.org/10.1016/j.jpsychires.2018.07.012
Thase M., Parikh S., Rothschild A., Dunlop B., Debattista C., Conway C., Forester B., Mondimore F., Shelton R., Macaluso M., Li J., Brown K., Jablonski M., and Greden J., 2019, Impact of pharmacogenomics on clinical outcomes for patients taking medications with gene-drug interactions in a randomized controlled trial, The Journal of Clinical Psychiatry, 80: 6.
https://doi.org/10.4088/JCP.19m12910
Tuson M., Pérez V., Salavert A., Mazo J., and Menchón J., 2017, Pharmacogenomic Information in the treatment of major depression: results from a randomized clinical trial, European Neuropsychopharmacology, 27: S364-S365.
https://doi.org/10.1016/j.euroneuro.2016.09.388
Wray N., Ripke S., Mattheisen M., Trzaskowski M., Byrne E., Abdellaoui A., Adams M., Agerbo E., Air T., Andlauer T., Bacanu S., Bækvad-Hansen M., Beekman A., Bigdeli T., Binder E., Blackwood D., Bryois J., Buttenschøn H., Bybjerg-Grauholm J., Cai N., Castelao E., Christensen J., Clarke T., Coleman J., Colodro-Conde L., Couvy-Duchesne B., Craddock N., Crawford G., Crowley C., Dashti H., Davies G., Deary I., Degenhardt F., Derks E., Direk N., Dolan C., Dunn E., Eley T., Eriksson N., Escott-Price V., Kiadeh F., Finucane H., Forstner A., Frank J., Gaspar H., Gill M., Giusti-Rodríguez P., Goes F., Gordon S., Grove J., Hall L., Hannon E., Hansen C., Hansen T., Herms S., Hickie I., Hoffmann P., Homuth G., Horn C., Hottenga J., Hougaard D., Hu M., Hyde C., Ising M., Jansen R., Jin F., Jorgenson E., Knowles J., Kohane I., Kraft J., Kretzschmar W., Krogh J., Kutalik Z., Lane J., Li Y., Li Y., Lind P., Liu X., Lu L., Macintyre D., MacKinnon D., Maier R., Maier W., Marchini J., Mbarek H., McGrath P., McGuffin P., Medland S., Mehta D., Middeldorp C., Mihailov E., Milaneschi Y., Milani L., Mill J., Mondimore F., Montgomery G., Mostafavi S., Mullins N., Nauck M., Ng B., Nivard M., Nyholt D., O'Reilly P., Oskarsson H., Owen M., Painter J., Pedersen C., Pedersen M., Peterson R., Pettersson E., Peyrot W., Pistis G., Posthuma D., Purcell S., Quiroz J., Qvist P., Rice J., Riley B., Rivera M., Mirza S., Saxena R., Schoevers R., Schulte E., Shen L., Shi J., Shyn S., Sigurdsson E., Sinnamon G., Smit J., Smith D., Stefánsson H., Steinberg S., Stockmeier C., Streit F., Strohmaier J., Tansey K., Teismann H., Teumer A., Thompson W., Thomson P., Thorgeirsson T., Tian C., Traylor M., Treutlein J., Trubetskoy V., Uitterlinden A., Umbricht D., Auwera S., Hemert A., Viktorin A., Visscher P., Wang Y., Webb B., Weinsheimer S., Wellmann J., Willemsen G., Witt S., Wu Y., Xi H., Yang J., Zhang F., Arolt V., Baune B., Berger K., Boomsma D., Cichon S., Dannlowski U., Geus E., DePaulo J., Domenici E., Domschke K., Esko T., Grabe H., Hamilton S., Hayward C., Heath A., Hinds D., Kendler K., Kloiber S., Lewis G., Li Q., Lucae S., Madden P., Magnusson P., Martin N., McIntosh A., Metspalu A., Mors O., Mortensen P., Müller-Myhsok B., Nordentoft M., Nöthen M., O'Donovan M., Paciga S., Pedersen N., Penninx B., Perlis R., Porteous D., Potash J., Preisig M., Rietschel M., Schaefer C., Schulze T., Smoller J., Stefánsson K., Tiemeier H., Uher R., Völzke H., Weissman M., Werge T., Winslow A., Lewis C., Levinson D., Breen G., Børglum A., and Sullivan P., 2017, Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression, Nature Genetics, 50: 668-681.
Westrhenen R., Aitchison K., Ingelman-Sundberg M., and Jukic M., 2020, Pharmacogenomics of antidepressant and antipsychotic treatment: how far have we got and where are we going? Frontiers in Psychiatry, 11: 94.
https://doi.org/10.3389/fpsyt.2020.00094
Zhou J.Y., 2024, The Application of genomics in personalized cancer therapy, Cancer Genetics and Epigenetics, 12(1): 66-74.
https://doi.org/10.5376/cge.2024.12.0009
(1).png)
. FPDF(win)
. FPDF(mac)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Tiantian Li
. Jie Zhang
Related articles
. Hepatitis B virus (HBV)
. Vaccination
. Epidemiology
. Hepatocellular carcinoma (HCC)
. Global health strategies
Tools
. Post a comment